Abstract
Abnormal degradation of matrix components due to dysregulated expression of matrix metalloproteinase (MMP)-9 in macrophages has been linked to progression of acute cerebral ischemia and atherosclerosis. We report that lithium chloride (LiCl) or CHIR99021, inhibitors of Wnt signaling pathway, enhance phosphorylation of glycogen synthase kinase-3beta and suppress lipopolysaccharide-mediated upregulation of MMP-9 expression in murine macrophage RAW264.7 cells in a dose- dependent manner. Suppression of MMP-9 expression by LiCl or CHIR99021 did not result after inhibition of kinases involved in NFkB or AP-1 family proteins, but from changes in the activity of histone deacetylases. Beneficial effects on atherosclerosis or cerebral ischemia in animal studies caused by LiCl may be in part explained by the suppression of MMP-9 gene expression.
Keywords: GSK-3b; HDAC; lipopolysaccharide; MMP-9; macrophage; MMP-9
Introduction
Dysregulated degradation of matrix components by matrix metalloproteinases (MMPs) has been linked to progression of several inflammatory diseases, such as cerebral ischemia, chronic obstructive pulmonary disease, and atherosclerosis (Rouis, 2005; del Zoppo et al., 2012). Expression levels and activity of MMP-9 are upregulated by inflammatory cytokines, hormones, or lipopolysaccharide (LPS) (Rouis, 2005). LPS interacts with the CD14, a main receptor for LPS, in conjunction with LPS-binding protein and the resulting LPS/CD14 complex interacts with Toll-like receptor 4 (TLR4) to activate a host of kinases in the down-stream signaling pathways. The LPS/CD14/TLR4 complex induces phosphorylation of IkB kinases and mitogen activating protein (MAP) kinases, which results in activation of transcription factors NFkB and AP-1 (Mackman et al., 1991; Lin et al., 2008).
MMP-9 expression can be regulated at the transcriptional or posttranscriptional levels. Transcription of MMP-9 is primarily upregulated through activation of transcription factors AP-1, NFkB, Sp1, and Ets1, which binds to their cis- elements on the MMP-9 promoter (Masure et al., 1993; Lin et al., 2008; Erdozain et al., 2011). Interaction of these transcription factors to the MMP-9 promoter requires chromatin remodeling factors, such as histone acetyl transferases (HATs) and histone deacetylases (HDACs), to induce transcription of MMP-9 (Ma et al., 2004; Zhao and Benveniste, 2008). Chromatin immunoprecipitation assays demonstrated that recruitment of HDAC-1 and -3, but not HDAC-2 and -4, suppressed MMP-9 gene expression (Ma et al., 2004).
In vitro and in vivo studies demonstrate that HDAC inhibitors suppress inflammatory genes, such as interleukin (IL)-1b, inducible nitric oxide synthase and tumor necrosis factor a (Adcock, 2007). HDACs regulate transcriptional efficiency by chromatin remodeling with deacetylation in core histones as well as in nonhistone transcription factors whose activities are markedly influenced by a degree of acetylation (Saha and Pahan, 2006). HDAC inhibitors have been intensively developed in hope to treat various types of inflammatory diseases (Adcock, 2007). A reduced form of nicotinamide adenine dinucleotide phosphate (NADP), but not NADP+, enhances HDAC activity (Vogelauer et al., 2012).
Our previous study demonstrated strong MMP-9 activity from the macrophages in and around “rupture-prone” inflammatory atherosclerotic plaques (Kim et al., 2010). In animals lithium chloride (LiCl), which phosphorylates and inhibits glycogen synthase kinase (GSK)-3b, alleviated atherosclerotic lesion formation in ApoE—/— mice fed a high-cholesterol diet (Choi et al., 2010). GSK-3b, whose activity is inhibited by phosphorylation on a serine residue 9, regulates activity of transcription factors AP-1, cAMP response element-binding protein, and NFkB, resulting in modulation of inflammatory mediators (Cortes-Vieyra et al., 2010). Phosphatidylinositide 3-kinases-Akt-depen- dent inhibition of GSK-3b has been reported to suppress production of pro-inflammatory cytokines (Wang et al., 2011). These led us: (1) to hypothesize that phosphorylation of GSK-3b would suppress upregulation of MMP-9 in macrophage upon LPS challenge, and (2) to investigate molecular mechanisms by which phosphorylation of GSK- 3b suppresses MMP-9 gene expression in macrophages. We report that LiCl and CHIR99021, GSK-3b inhibitors, suppress LPS-mediated upregulation of MMP-9 expression by modulating HDACs activity in murine macrophage RAW264. cells.
Materials and methods
Materials
Atorvastatin, LiCl, LPS from Escherichia coli 0111:B4, NADPH, trichostatin A (TSA), and simvastatin were purchased from Sigma–Aldrich (St. Louis, MO). CHIR99021, a GSK-3b inhibitor, and (2’Z,3’E)-6-bromoin- dirubin-3′-oxime (BIO), a highly selective GSK-3 inhibitor IX, were purchased from Calbiochem (Darmstadt, Germany). CHIR99021, simvastatin, BIO, and TSA were dissolved in dimethylsulfoxide (DMSO) and diluted with cell culture medium immediately before adding to cell culture medium. Primary antibodies against p38, phosphorylated p38, ERK 1/2, phosphorylated ERK 1/2, JNK 1/2, phos- phorylated JNK 1/2 which is also known as stress-activated protein kinases, GSK-3b phosphorylated at a serine residue 9 or GSK-3a phosphorylated a serine residue 27, acetylated- lysine, and b-tubulin were purchased from Cell Signaling
Technology (Danvers, MA). FastStart PCR Master Mixes and phosphatase inhibitor cocktail were purchased from Roche Applied Sciences (Mannheim, Germany).
Cell culture
RAW264.7 murine macrophages were purchased from Korean Cell Line Bank (Seoul, Korea) and maintained in phenol red free DMEM supplemented with 10% heat- inactivated fetal bovine serum, 100 U/mL penicillin, and 100 mg/mL streptomycin at 37◦C in a humidified incubator containing 5% CO2. Cells used in all experiments were subjected to no higher than 27 cell passage numbers. When cells in 12-well plates reached ~80% confluency, they were washed with PBS, incubated in serum-free DMEM, and treated with 0.1 mg/mL LPS from E. coli 0111:B4 and vehicle (DMSO) or various reagents for 10 h or as indicated. Final concentrations of DMSO in all cell culture experiments were adjusted to 0.05%.
Synthesis of cDNA and quantitative real-time polymerase chain reaction (RT-PCR)
Medium was collected for zymography, and total RNA was extracted with TRIsure (Bioline). Synthesis of cDNA was performed using Quantitect Reverse Transcription Kit (Qiagen) according to the manufacturer’s instruction. Analy- sis of mRNA expression was determined with quantitative real-time PCR using 10 pmol primers of b-actin (forward- 5’CTAAGGCCAACCGTGAAAAG3′ reverse-5’ACCAGA- GGCATACAGGGACA3′) as a reference gene and 10 pmol primers of MMP-9 (forward-5’AAACCAGACCCCAG- ACTCCTC3′ reverse-5’GAGGACACAGTCTGACCTGA-A3′). Abundance of MMP-9 mRNA in each sample was determined by the DCt (cycle threshold), the difference between the Ct values for MMP-9 and an internal control gene b-actin. Relative ratios of MMP-9 mRNA expression levels were defined as 2—DDCt which reflects changes of DCt of cells with LPS treatment compared to DCt of unstimulated cells. Ratios of MMP-9 mRNA to b-actin mRNA in the presence of LPS without LiCl or CHIR99021 were calculated as 1 in each set of experiment for statistical analysis. Statistical analyses were determined from at least three independent experiments.
Gelatin zymography
Aliquots of medium mixed with zymography sample buffer (Bio-Rad) were incubated for 1 h at room temperature and analyzed by SDS–PAGE with 10% gels containing 1 mg/mL gelatin at 4◦C. Proteins in the gels were renatured by incubating for 30 min at 37◦C in 50 mM Tris–HCl/pH 7.4, 2.5% Triton X-100, 5 mM CaCl2, and 1 mM ZnCl2. The gels were incubated in 50 mM Tris–HCl/pH 7.4, 5 mM CaCl2, and 1 mM ZnCl2 overnight at 37◦C. The gels were stained with 0.25% (w/v) Coomassie Brilliant Blue R250 solution in 20% methanol and 5% acetic acid, and destained in 20% methanol and 5% acetic acid.
Flow cytometry for analysis of CD14 expression levels
Cells treated with 5 mM LiCl and/or LPS (0.1 mg/mL) for 3 or 20 h were collected, washed with PBS, labeled with 3% normal mouse serum and anti-CD14-FITC (BD Pharmingen) in FACS buffer (0.1% BSA and 0.02% sodium azide in PBS) for 30 min at 4◦C, washed with PBS twice, fixed with 4% paraformaldehyde in PBS, washed with PBS, resus- pended in FACS buffer, and analyzed by Beckman Coulter flow cytometer. A sample of cells incubated with FITC- conjugated nonspecific isotype antibody served as a negative control.
Preparation of cell lysates and Western blotting analysis
Cells in serum-free DMEM were treated with ligands (vehicle, LiCl, or CHIR) and/or LPS for 40 min or otherwise indicated, lysed by adding high salt cell lysis buffer (20 mM Tris–HCl/pH 7.5, 1 mM EDTA, 1 mM EGTA, 1% Triton X- 100, 1 mg/mL leupeptin, 2.5 mM sodium pyrophosphate, 1 mM b-glycerophosphate, 1 mM Na3VO4, 0.3 M NaCl,0.5 mM phenylmethanesulfonyl fluoride) plus phosphatase inhibitor cocktail, and centrifuged at 12,000g for 5 min at 4◦C. Proteins in total cell lysates were quantitated by a Bradford assay and separated by SDS–PAGE, and analyzed by Western blot using enhanced chemiluminescence detection kit.
Cell viability and proliferation assays
Cells were stained by adding an equal volume of 0.4% trypan blue in PBS. Portions of dead cells, stained by trypan blue, in a given field under the microscope were calculated. Cell proliferation rates were determined by MTT (3-(4,5- dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, a tetrazole) assay. MTT solution (final concentration 0.4 mg/ mL) was added to cells grown in phenol red free medium, and cells were incubated for 3 h. Medium was removed, and the converted formazan dye was solubilized with acidic isopropanol (10 mM HCl in absolute isopropanol). Absor- bance at 570 nm with background subtraction at 650 nm was measured.
Statistical analysis
Data are presented as mean standard deviation (SD). Statistical comparison of data was determined using one-way analysis of variance, followed by the Tukey–Dunnett post hoc adjustment. P < 0.05 was considered significant.
Results
LiCl and CHIR99021 suppress LPS-mediated upregulation of MMP-9 gene expression in a dose- dependent manner
LPS induced transcriptional activation of MMP-9 in murine macrophage RAW264.7 cells, as judged by a quantitative real-time RT-PCR and zymography assays. Treatment of cells with different concentrations of LiCl effectively suppressed LPS-mediated transcriptional activation of MMP-9 in a dose-dependent manner (Figure 1A). CHIR99021, a highly selective GSK-3b inhibitor, also suppressed LPS-induced MMP-9 expression in a dose- dependent manner (Figure 1B). Treatment of cells with LiCl or CHIR99021 after 1 h of LPS treatment suppressed LPS- induced transcriptional activation of MMP-9 (data not shown), indicating that prior treatment of cells with LiCl or CHIR99021 was not required for suppression of LPS- mediated MMP-9 gene expression. LiCl or CHIR99021 did not show statistically significant changes in cell viability or a proliferation rate under the concentrations used (data not shown), demonstrating that suppression of LPS-mediated MMP-9 gene expression did not result from a change of cell viability after treatment of LiCl or CHIR99021.
Atorvastatin and simvastatin (5 mM) enhance LPS-mediat- ed MMP-9 gene expression in RAW264.7 (Lee et al., 2012). LiCl and CHIR99021 effectively suppressed the upregulation of atorvastatin or simvastatin-mediated upregulation of LPS- induced MMP-9 expression(Figure 2A). Inadditionto MMP- 9, LPS upregulates inflammatory cytokines in macrophages. We determined whether LiCl suppresses LPS-induced interleukin (IL)-1b and IL-6 gene expression in RAW264.7 cells. LPS markedly enhanced mRNA expression levels of IL- 1b and IL-6 (Figures 2B and 2C). LiCl suppressed LPS- induced mRNA expression of IL-1b and IL-6 genes in a dose- dependent manner (Figures 2B and 2C). However, LiCl did not suppress LPS-induced expression of induced nitric oxide in RAW264.7 cells (data not shown).
Since activity of GSK-3 is suppressed by phosphorylation of Ser9 (GSK-3b) or Ser21 (GSK-3a), phosphorylation patterns of GSK-3 were analyzed by a Western blot assay. LiCl and CHIR99021 preferentially enhanced phosphoryla- tion of Ser9 in GSK-3b (Figure 3A). A highly selective GSK- 3b inhibitor BIO, which could enhance phosphorylation of GSK-3b (Figure 3A), was used to confirm that inhibition of GSK-3b would effectively suppress the LPS-mediated MMP- 9 gene expression in macrophages. GSK-3b inhibitor BIO suppressed LPS-mediated MMP-9 gene expression at a micromolar concentration (Figure 3B).
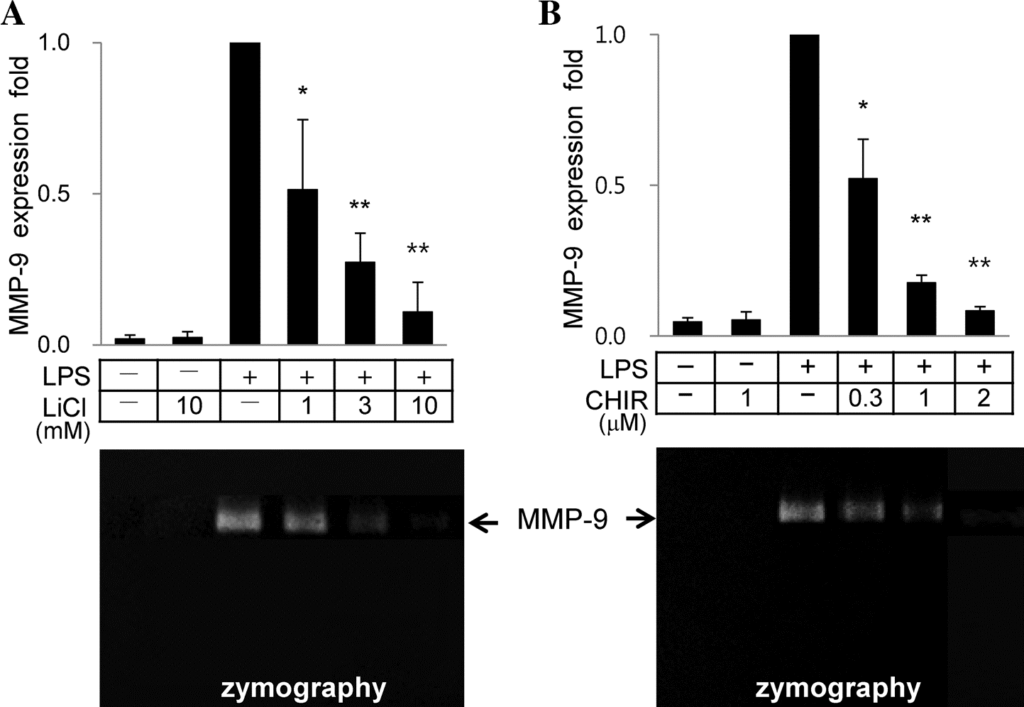
Figure 1 LiCl and CHIR99021 suppress LPS-induced gene expression of MMP-9 in a dose dependent manner. RAW264.7 cells were treated with LPS (0.1 mg/mL) and/or different concentrations of LiCl (A) or CHIR99021 (B). Real-time RT-PCR of MMP-9 was carried out with b-actin as an internal control. Ratios of MMP-9 mRNA to b-actin mRNA in the presence of LPS without LiCl or CHIR99021 were calculated as 1 in each set of experiments for statistical analysis. Results are presented as means SD; P < 0.05 versus LPS only.P < 0.01 versus LPS only.
LiCl and CHIR99021 neither suppress expression levels of CD14 nor phosphorylation efficiency of kinases involved in activation of transcription factors NFkB or AP-1
We determined if LiCl-mediated suppression of MMP-9 expression resulted from reduced cell surface expression levels of CD14. Expression levels of CD14 were upregulated upon LPS challenge in two steps, the first rapid increase in 30–180 min by translocation of intracellular CD14 pool to the plasma membrane and the second upregulation associated with de novo protein synthesis of CD14 (Antal- Szalmas, 2000). FACS analysis showed that LPS treatment increased cell surface expression of CD14 (Figure 4A) as early as 3 h after LPS treatment. However, LiCl did not change cell surface expression of CD14 at 3 h (LPS: 9.8 2.15% vs. LPS/LiCl: 9.5 1.87%) or 20 h (LPS: 71.2 4.63% vs. LPS/LiCl 70.7 4.81%) after LPS challenge, indicating that LiCl changed neither intracellular trans- location of CD14 nor de novo protein synthesis of CD14 (Figure 4A). No change in cell surface expression pattern of CD14 was also observed with CHIR99021 (data not shown).
We analyzed if LiCl or CHIR9902 inhibited phosphoryla- tion efficiency of kinases involved in the LPS/CD14/TLR4 complex-mediated signaling pathways. As reported before (Woo et al., 2008; Burk et al., 2009), treatment of cells with LPS resulted in phosphorylation and activation of the MAP kinases p38, Erk1/2, and JNK/SAPK (Figure 4B). However, treatment of cells with LiCl did not decrease phosphoryla- tion efficiency of p38, Erk1/2, or JNK/SAPK in the presence of LPS (Figure 4B). Treatment of cells with LPS increased phosphorylated form of IkB and decreased an amount of total IkB in cells. However, LiCl did not decrease the phosphorylated form of IkB (Figure 4B). Indeed, LiCl did not change the degree of nuclear translocalization of NFkB. Treatment of cells with CHIR99021 did not suppress phosphorylation of p38, ERK1/2, and JNK (data not shown). LiCl or CHIR99021 did not suppress LPS-induced MMP-9 gene expression by enhancing activity of AP-1 family proteins or NFkB (Figure 4).
Histone deacetylation/acetylation activity may play a crucial role in suppression of LPS-mediated MMP-9 gene expression by LiCl or CHIR99021
We asked whether alteration of the activity of HDACs that could affect access of transcription factors and subsequently change gene expression (Adcock, 2007; Dokmanovic et al., 2007), were necessary for suppression of LPS-mediated MMP-9 gene expression by LiCl or CHIR99021. Since HDACs change acetylation patterns of lysine residues in histone as well as nonhistone proteins and modulate gene expression, we determined if LiCl or CHIR99021 changed acetylation patterns of lysine residues of the nuclear proteins. Nuclear extracts obtained from cells treated with LiCl, CHIR99021, or BIO were analyzed using an antibody against acetylated lysine residue. While LPS alone did not change acetylation patterns of nuclear proteins, treatment of cells with LiCl, CHIR99021, or BIO enhanced the intensity of an acetylated protein with an apparent molecular weight of 60 and decreased that with an apparent molecular weight of 33 kDa (Figure 5A). It is unclear how acetylated proteins with apparent molecular weights of 60 and 33 kDa are involved in MMP-9 expression.
Figure 2 LiCl suppresses upregulation of LPS-mediated MMP-9, IL- 1b, and IL-6 expression. (A) Cells were treated with LPS (0.1 mg/mL) and vehicle or ligands (5 mM LiCl, 1 mM CHIR99021, 5 mM atorvastatin or simvastatin) as indicated for 8 h and subjected to RNA isolation. Real-time RT-PCR of MMP-9 was carried out as described in Figure 1. (B,C) Cells were treated with LPS (0.1 mg/mL) and different concentrations of LiCl for 8 h and subjected to RNA isolation. Real-time RT-PCR of IL-1b or IL-6 was carried out as described in Figure 1. Results are presented as means SD.P < 0.05. P < 0.01.
TSA, a highly potent inhibitor for HDAC, decreased LPS- mediated MMP-9 gene expression (Figure 4B). However, TSA showed little effect in decreasing LiCl or CHIR- mediated suppression of LPS-induced MMP-9 expression (indicated by # in Figure 5B). We further analyzed if activation of HDACs could reverse suppression of LPS- mediated MMP-9 gene expression by LiCl or CHIR99021. Addition of NADPH, which can activate HDACs (Vogelauer et al., 2012), alleviated suppression of LPS-mediated MMP-9 expression by LiCl or CHIR99021 (Figure 5C). LiCl or CHIR99021 might change acetylation of lysine residues in certain nuclear proteins and suppress LPS-mediated MMP-9 gene expression (Figure 4).
Discussion
Dysregulated upregulation of inflammatory cytokines as well as MMP-9 has been associated with progression of inflammatory diseases, such as atherosclerosis, cerebral ischemia, and sepsis (Lorente et al., 2009). Lithium ion, which phosphorylates GSK-3 and inhibits the GSK-3 signaling pathway, has been reported to exert pleiotropic effects on several inflammatory diseases (Choi et al., 2010; Albayrak et al., 2013). We report here that LiCl suppressed LPS-induced MMP-9 gene expression in murine macro- phage RAW264.7 cells in a dose-dependent manner. LiCl also suppressed LPS-induced IL-1b and IL-6 gene expression. Along with LiCl, other GSK-3 inhibitors including CHIR99021 and BIO also suppressed LPS-induced MMP-9 gene expression in RAW264.7 cells.
Figure 3 BIO, a GSK-3b inhibitor, suppresses LPS-induced MMP-9 gene expression in murine macrophages. (A) Cells were treated with LPS (0.1 mg/mL) and ligands (5 mM LiCl, 1 mM CHIR99021, or 1 mM BIO) as indicated for 3 h, and cell lysates were analyzed by Western blot with antibody against phosphorylated GSK-3. (B) Cells were treated as indicated for 10 h, total RNA was isolated, and real-time RT-PCR of MMP-9 was carried out as described in Figure 1. Results are presented as means SD; P < 0.05 versus LPS only.
Figure 4 LiCl neither suppresses expression levels of CD14 nor phosphorylation efficiency of kinases involved in activation of transcription factors NFkB or AP-1. (A) LiCl does not change surface expression levels of CD14. Cells were treated with 5 mM LiCl and/or LPS (0.1 mg/mL) for 3 or 20 h, labeled with anti-CD14-FITC, and analyzed by flow cytometer. A sample of cells incubated with FITC-conjugated nonspecific isotype antibody served as a negative control. (B) LiCl or CHIR99021 does not change phosphorylation efficiency of kinases in the MAPK and NFkB pathways. RAW264.7 cells in serum-free medium were stimulated with LPS in the presence or absence of 5 mM LiCl, and cell lysates were analyzed by Western blot with antibodies against p38, phosphorylated form of p38 (P-p38), Erk 1/2, phosphorylated form of Erk 1/2 (P-Erk 1/2), JNK/SAPK, phosphorylated form of JNK/ SAPK (P-JNK), IkB or phosphorylated form of IkB.
A series of biological alterations in the macrophages following formation of the LPS/CD14/TLR4 complex are mainly due to the activation of transcription factors NF- kB and AP-1. A number of plant extracts, such as epigallocatechin gallate from green tea and curcumin from curry, suppress NFkB and/or AP-1 by changing phosphorylation efficiency of IkB and/or kinases in the MAP kinase signaling pathway (Kim et al., 2005; Burk et al., 2009; Hong Byun et al., 2010). However, we found that LiCl and CHIR99021 downregulated LPS-induced MMP-9 gene expression without suppressing activity of AP-1 or NFkB.
Figure 5 LiCl and CHIR99021 suppress LPS-induced MMP-9 gene expression by modulating HDAC activity. (A) Cells were treated with LPS (0.1 mg/mL) and ligands (5 mM LiCl, 1 mM CHIR99021, 2 mM BIO) as indicated for 4 h and subjected for preparation of whole cell lysates. Whole cell lysates were separated and analyzed with antibody against acetylated-lysine. The arrows indicate two proteins with changes in acetylation patterns by treatment with LiCl, CHIR, or BIO. (B) Cells were treated with 5 mM LiCl, 1 mM CHIR99021, or 50 ng/mL TSA for 8 h and subjected for total RNA isolation.(C) Cells were treated with LiCl, CHIR99021, or 0.1 mM NADPH for 8 h and subjected for total RNA isolation. Real-time RT-PCR of MMP-9 was carried out as described in Figure 1. Results are presented as means SD. P > 0.05, differences are not statistically significant. P < 0.05, differences are statistically significant.
Alteration of mRNA stability is another step to regulate MMP-9 gene expression. Nitric oxide suppresses the expression of HuR, an mRNA stabilizing factor interacting with the AU-rich elements in the 3′ untranslated region, and increases the decay rate of MMP-9 mRNA in glomerular mesangial cells (Akool el et al., 2003). Others have reported that LiCl increases the decay rate of insulin receptor mRNA~50% in adrenal chromaffin cells (Yokoo et al., 2007). However, LiCl and CHIR99021 did not suppress LPS- mediated MMP-9 gene expression by increasing a decay rate of MMP-9 mRNA (data not shown). Further studies are required to elucidate the differential effects of LiCl on the stability of MMP-9 mRNAs and insulin receptor mRNAs.
Changes in chromatin structure caused by acetylation of core histones transform condensed chromatin into a relaxed structure which allows access of transcription factors (Saha and Pahan, 2006). In contrast, recruitment of HDACs to chromatin can cause condensation of chroma- tin structure and gene silencing (Li and Greene, 2007). However, inhibition of histone deacetylation does not cause gene expression of every gene, as certain genes are induced while others are suppressed by HDAC inhibitors (Kim et al., 2001; Nair et al., 2001). The complex effects of HATs and HDACs on patterns of gene expression are in part due to the fact that HATs and HDACs participate in acetylation and deacetylation of nonhistone transcription factors whose activities are markedly influenced by a degree of acetylation (Saha and Pahan, 2006). Biochemical and molecular analyses of MMP-9 gene expression in HeLa cells using chromatin immunoprecipitation assay demonstrated that HDAC-1 and HDAC-3 were released from the MMP-9 promoter region upon activation by phorbol 12- myristate 12-acetate (Ma et al., 2004).
TSA belongs to a class I HDAC inhibitor which blocks activity of HDAC-1, HDAC-2, HDAC-3, and HDAC-8 (Dokmanovic et al., 2007).The monovalent cation lithium in the chloride or carbonate form has been used to treat patients with bipolar disorder for 50 years (Thakker-Varia et al., 2010). Apart from the beneficial effect of LiCl as a mood stabilizer, the protective roles of LiCl in atherosclerotic lesion formation have been explored (Choi et al., 2010).
LiCl also suppresses the expression of pro-inflammatory cytokines due to bacteria-induced inflammation (Duan et al., 2007). We report that LiCl and CHIR99021 effectively suppress MMP- 9 gene expression, which can provide another mechanism to explain beneficial roles of LiCl to interfere with progress of atherosclerosis. Given the evidence indicating that expres- sion of MMP-9 is upregulated in vulnerable rupture-prone atherosclerotic plaques (Papaspyridonos et al., 2006; Kimet al., 2010), LiCl poses an attractive strategy to suppress an expression level of MMP-9 and prevent rupture of vulnerable plaques.